
Fast deconvolution of 3D Light-Sheet-Images
VALID combines theoretical considerations and the usage of empiric image data to alculate the true dimensions of the a light sheet. This allows to deconvolve a 3D image stack by a precise subtraction of the bleed-through signal between the single slices at each position of the light sheet in 3D. Furthermore, VALID extracts information allowing to define the optimal distaince between the single slices during imaging, which reduces data size and speeds up imaging itself, as well as all subsequent image processing. This approach is surperior to known deconvolutuion methods, which are either made for pointwise (e.g. confocal) imaging or use crude estimations for the 3D shape of the lightsheet.
Challenge
Light Sheet Microscopy (LSM) offers a unique combination of high frame rate and good resolution for large samples. Large tissues and whole organisms can be observed at cellular resolution, which provides information that cannot be imaged with other methods. In classical Microscopy, deconvolution is a standard method to improve images and extract even more information. However, classical deconvolution models are not suitable, are too slow for the unfolding of 3D light sheet microscopic images. A deconvolution software for 3D image stacks is highly desired to achieve enhancement of information.
Our Solution
Inventors at the University Medical Center Göttingen have developed a deconvolution method that improves image quality in all three dimensions of Light Sheet Images - fast and reliable. The development of a combined empirical and theoretical approach using light sheet image data from a microscope makes it possible to calculate the real shape of the lightsheet. But also additional properties of the sample and the light sheet can be extracted from the measurement. This allows to reduce the
number of slices for a 3D image, since the optimal distance between the single steps in z-direction can be calculated and used for imaging. Compared to using the Nyquist criteria, the number of z-steps can be significantly reduced, resulting in faster imaging and less data - without loosing any information.
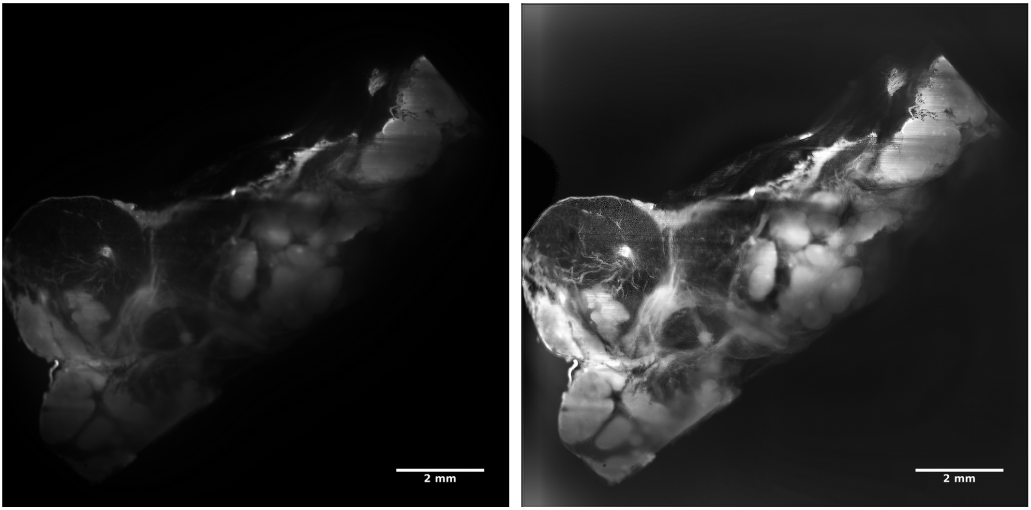 Figure 1: Human thymus, cleared, autofluorescence. Tissue depth: 3,3 mm.
Figure 1: Human thymus, cleared, autofluorescence. Tissue depth: 3,3 mm.
Left: before deconvolution. Right: after deconvolution
For the deconvolution of a complete image stack of 600 images with 2048x2048 pixels, about 8 hours were required using the Richardson-Lucy algorithm and locally variable psf. The deconvolution with the newly developed method was completed on the same conmputer in approximately 5 minutes.
Advantages
- deconvolution is based on the real lightsheet shape
- fast and reliable deconvolution signficantly improves image quality and reveals more details
- optimization of step width is possible without losing information
- decreases the number of image slices - reducing data
- allows faster imaging with lower overall light dose
- allows fast and robust deconvolution
- The algorithm can be applied to other tomographich imaginging methods like e.g. MRI
Applications
- Deconvolution of 3D stacks from light sheet images
Development Status
The technology is used routinely in the lab of the inventors.
Patent Status
A german priority patent application and am interlational PCT patent application are pending, which enables us to provide a licensee with worldwide protection rigths. The international search report granted novelty and inventiveness to the core of the invention.
References
Pending PCT-Patent Application: WO2021074213A1
Contact
Dr. Martin Andresen
Patent Manager Life Sciences
E-Mail: This email address is being protected from spambots. You need JavaScript enabled to view it.
Tel.: +49 551 30724 150
Reference: CPA-2168-UMG
Tags: MRI, Computer tomography, 3D Imaging, Deconvolution, Light Sheet Imaging
