Method to visualize proteins and protein-complexes with sizes of less than 1nm
Researchers in the Rizzoli Lab at the university Göttingen developed a method which allows to visualize protein-protein interactions and might even be suitable to resolve 3D protein structures. Combining expansion microscopy with "super-resolution radial fluctuations" (SRRF) allows to reach resolutions in the sub nm range - in principle. The inventors finally succeeded to take images of single proteins by the development of an improved sample preparation, well defined imaging parameters and a smart image processing. The method can be used with comparably simple and cheap fluorescence microscopes and may allow scientists all over the world to investigate the world of protein interactions without spending millions of Euros for high-end hardware and weeks or months of work for a single image.
Challenge
Optical microscopy is one of the most valuable tools in understanding the functioning of organisms on cellular and sub-cellular levels for more than two centuries. The understanding of cellular processes was considerably enhanced by the introduction of high resolution methods like Laser Scanning Microscopy, followed by super-resolution microscopy allowing e.g. to study cellular substructures and the localization of proteins. But actually the basis of most biological processes, the interaction of proteins on a molecular level has hidden from light microscopy and stayed in the dark. It was necessary to use methods like NMR, X-ray crystallography or electron microscopy, which are very expensive and time consuming. Since many diseases are based on malfunctioning proteins or changes of protein expression, an A affordable way to visualize and study the shapes of single proteins and protein complexes may be a new breakthrough for understanding biological processes.
Our Solution
Scientists from the University of Göttingen found a solution to this problem, by combining Expansion Microscopy (ExM - a method allowing 10-fold axial expansion of the specimen - 1000-fold by volume) with a method named super-resolution radial fluctuations (SRRF). Additionally the inventors had to improve sample preparation, optimize imaging parameters and the image processing, since the microscopy method was not suitable for such high resolutions. The resulting Method is named Optimized Nanoscale Expansion Microscopy (ONE-Microscopy).
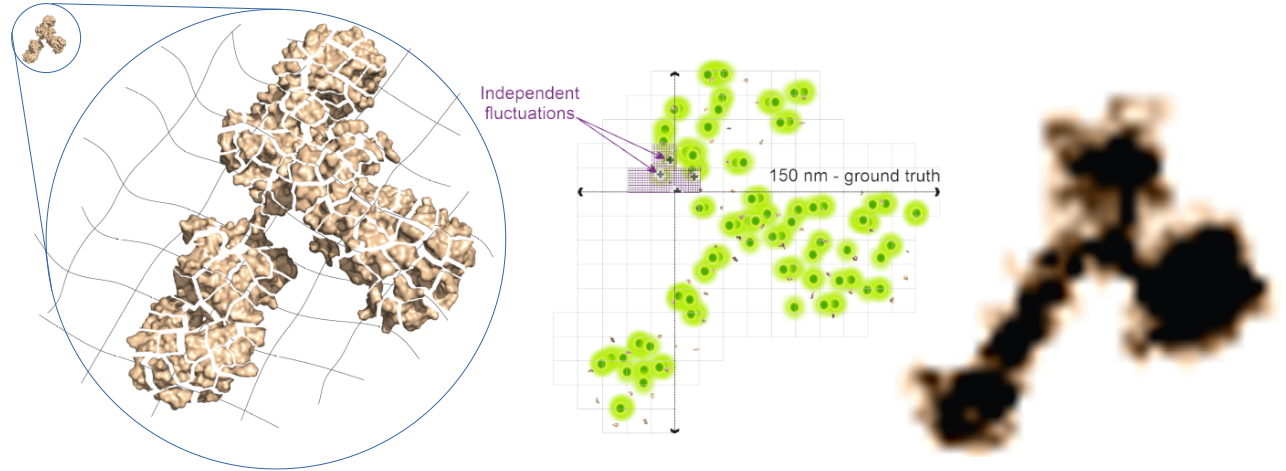 Fig. 1: Principle of ONE-Microscopy: Left: A labelled Protein (small Structure in the upper left corner) is anchored to an expandable Gel-Matrix, homogenized by treatment with Proteinase and Heat and then expanded with water by a factor of ~10. Middle: Imaging using SRRF. Right: Reconstructed 3D image of a single protein.
Fig. 1: Principle of ONE-Microscopy: Left: A labelled Protein (small Structure in the upper left corner) is anchored to an expandable Gel-Matrix, homogenized by treatment with Proteinase and Heat and then expanded with water by a factor of ~10. Middle: Imaging using SRRF. Right: Reconstructed 3D image of a single protein.
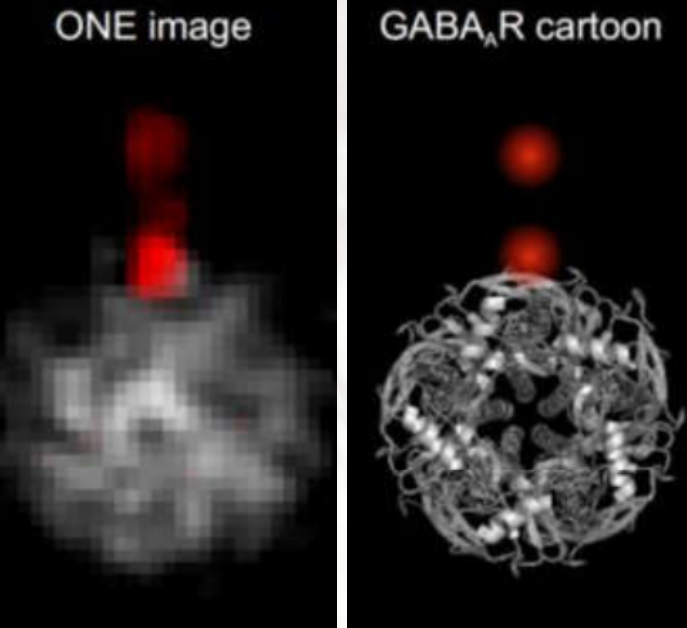
Using the rather cheap and simple ONE-Microscopy to image fluorescently labelled GABA-A Receptor Proteins it was possible to obtain 2D images of proteins, exhibiting details which are otherwise only possible to see with NMR or X-ray Crystallography (Figure2 left).
Furthermore it was possible to discriminate different shapes of alpha-synuclein oligerms and it was found that a bunch of seven oligmers turned out to be more frequent in Parkinsons Disease Patients (Figure2 mid). An overall score was developed, which allows to discriminate PD patients from non-PD-patients (Figure2 right).
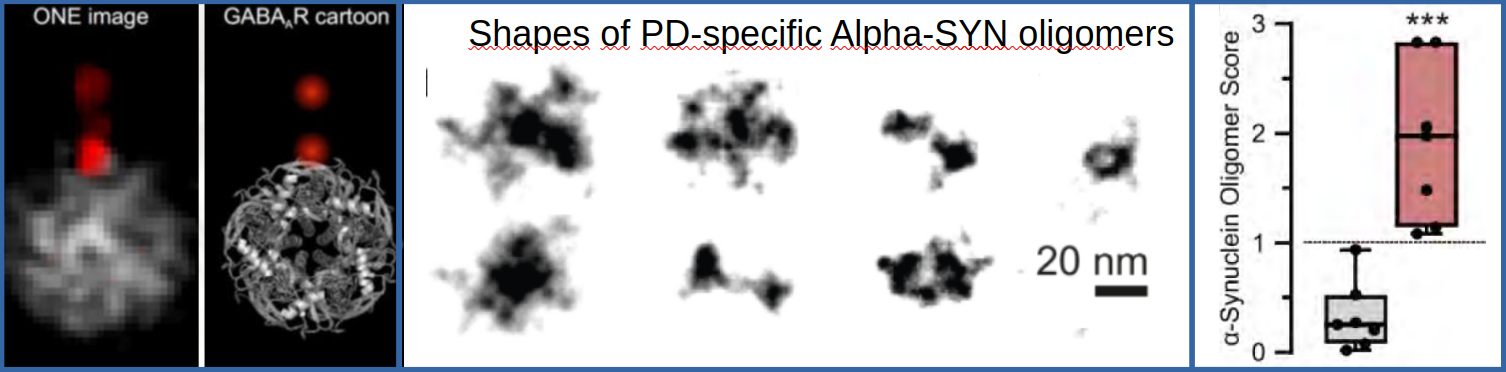 Fig. 2: Left: Comparison of averaged ONE-Images of the GABA-Receptor and a cartoon represntation of a PDB X-ray structure. Middle and right: Cerebro-spinal fluid probes were obtained from PD patiens and a control group. ONE-Imaging revealed a group of PD-specific ASYN-Cluster shapes. A comparison of the abundance of the different clusters in PD and control group yielded a score which seems to allow a sharp discrimination of PD patients from non-PD-patients.
Fig. 2: Left: Comparison of averaged ONE-Images of the GABA-Receptor and a cartoon represntation of a PDB X-ray structure. Middle and right: Cerebro-spinal fluid probes were obtained from PD patiens and a control group. ONE-Imaging revealed a group of PD-specific ASYN-Cluster shapes. A comparison of the abundance of the different clusters in PD and control group yielded a score which seems to allow a sharp discrimination of PD patients from non-PD-patients.
The technology has the potential to bring another revolution into the world of light microscopy in scientific research. It opens up the possibility to visualize protein shapes, conformational changes and protein interactions, which are in many cases altered in diseases. The ONE-Microscopy has the potential to identify many diseases resulting from malfunctioning proteins.
Advantages
- Resoultion of less than 1 nm
- flexible resolution depending on whether microscopic, nanoscopic or structure determination is needed
- visualization of the relative position of proteins in a complex
- No size restriction – even larger protein clusters can be analysed
- visualization of protein interactions or conformational changes
- simple, fast and cost effective 3D protein strucutre analysis may be possible
- Novel, scalable diagnostic approach to determine the origin of a disease
- Diagnosis of Parkinson Disease is possible in blood samples
- Diagnosis is cheap enough to be performed as standard in clinics
Applications
- Classical Imaging with flexible resolution
- Analysis of Protein function and Protein interaction
- Device for diagnosis of diseases on the protein-structure level like PD
- Structure aided drug design
- Identification and verification of drug targets
Development Status
- Proof of diagnostic principle with Parkinson Disease as an example
- Proof of concept for 3D Protein strucutre determination
Patent Status
Pending patent applications: PCT-patent application
Applicant: University Medical Center of Göttingen
Contact
Dr. Martin Andresen
Patent Manager Life Sciences
E-Mail: Diese E-Mail-Adresse ist vor Spambots geschützt! Zur Anzeige muss JavaScript eingeschaltet sein!
Tel.: +49 551 30724 150
Reference: BioT-2411-UMG
Tags: NMR, drug screening, A-SYN, alpha synuclein, ExM, Expansion Microscopy, SRRF, Protein Structure, 3D Protein structure, Parkinson Disease, diagniostic, structure determination, super resolution microscopy, ONE Microscopy, Imaging, Nanoscopy
